As part of daily routine, the laboratory microbiologist often has to determine the number of bacteria in a given sample as well as having to compare the amount of bacterial growth under various conditions. Enumeration of microorganisms is especially important in dairy microbiology, food microbiology, and water microbiology.
Since the enumeration of microorganisms involves the use of extremely small dilutions and extremely large numbers of cells, scientific notation is routinely used in calculations. A review of exponential numbers, scientific notation, and dilutions is found in Appendix B.
A. THE PLATE COUNT (VIABLE COUNT)
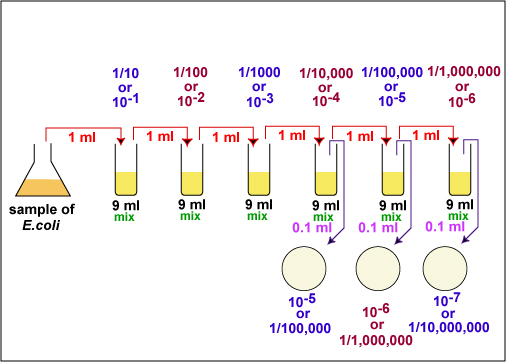
The number of bacteria in a given sample is usually too great to be counted directly. However, if the sample is serially diluted (see Fig. 1) and then plated out on an agar surface in such a manner that single isolated bacteria form visible isolated colonies (see Fig. 2), the number of colonies can be used as a measure of the number of viable (living) cells in that known dilution. However, keep in mind that if the organism normally forms multiple cell arrangements, such as chains, the colony-forming unit may consist of a chain of bacteria rather than a single bacterium. In addition, some of the bacteria may be clumped together. Therefore, when doing the plate count technique, we generally say we are determining the number of Colony-Forming Units (CFUs) in that known dilution. By extrapolation, this number can in turn be used to calculate the number of CFUs in the original sample.
Fig. 1: Plate Count Dilution Procedure
Fig. 2: Single Isolated Colonies Obtained During the Plate Count
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Normally, the bacterial sample is diluted by factors of 10 and plated on agar. After incubation, the number of colonies on a dilution plate showing between 30 and 300 colonies (see Fig. 2) is determined. A plate having 30-300 colonies is chosen because this range is considered statistically significant. If there are less than 30 colonies on the plate, small errors in dilution technique or the presence of a few contaminants will have a drastic effect on the final count. Likewise, if there are more than 300 colonies on the plate, there will be poor isolation and colonies will have grown together.
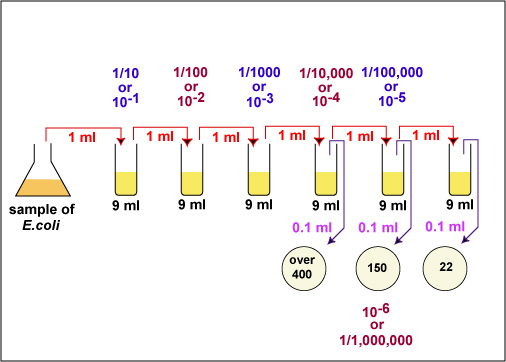
Generally, one wants to determine the number of CFUs per milliliter (ml) of sample. To find this, the number of colonies (on a plate having 30-300 colonies) is multiplied by the number of times the original ml of bacteria was diluted (the dilution factor of the plate counted). For example, if a plate containing a 1/1,000,000 dilution of the original ml of sample shows 150 colonies, then 150 represents 1/1,000,000 the number of CFUs present in the original ml. Therefore the number of CFUs per ml in the original sample is found by multiplying 150 x 1,000,000 as shown in the formula below:
The number of CFUs per ml of sample =
The number of colonies (30-300 plate) X
The dilution factor of the plate counted
In the case of the example above (see Fig. 3A and Fig. 3B), 150 x 1,000,000 = 150,000,000 CFUs per ml. (1.5 X 102 X 106 = 1.5 X 108 in scientific notation.)
Fig. 3A: Practice Plate Count Dilution Problem, Step 1
Fig. 3B: Practice Plate Count Dilution Problem, Step 2
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
For a more accurate count it is
advisable to plate each dilution in duplicate or triplicate and then find an
average count.
B. DIRECT MICROSCOPIC METHOD (TOTAL CELL COUNT)
In the direct microscopic count, a counting chamber consisting of a ruled slide and a coverslip is employed.. It is constructed in such a manner that the coverslip, slide, and ruled lines delimit a known volume. It is constructed in such a manner that the coverslip, slide, and ruled lines delimit a known volume. The number of bacteria in a small known volume is directly counted microscopically and the number of bacteria in the larger original sample is determined by extrapolation.
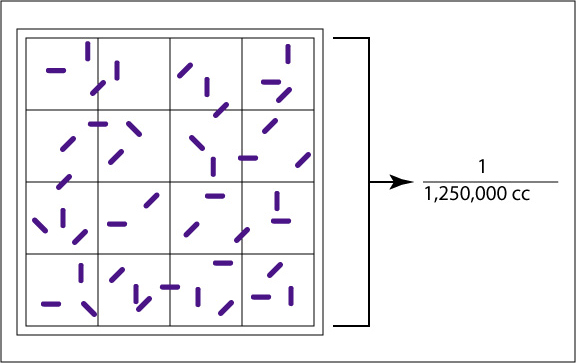
The Petroff-Hausser counting chamber (see Fig. 4A and Fig. 4B) for example, has small etched squares 1/20 of a millimeter (mm) by 1/20 of a mm and is 1/50 of a mm deep. The volume of one small square therefore is 1/20,000 of a cubic mm or 1/20,000,000 of a cubic centimeter (cc). There are 16 small squares in the large double-lined squares that are actually counted, making the volume of a large double-lined square 1/1,250,000 cc. The normal procedure is to count the number of bacteria in five large double-lined squares and divide by five to get the average number of bacteria per large square. This number is then multiplied by 1,250,000 since the square holds a volume of 1/1,250,000 cc, to find the total number of organisms per cc in the original sample.
Fig. 4A: Large Double-Lined Square of a Petroff-Hausser Counter
Fig. 4B: Petroff-Hausser Counter as seen through a Microscope
The large, double-lined square holds a volume of 1/1,250,000 of a cubic centimeter. Using a microscope, the bacteria in the large square are counted.
The double-lined "square" holding 1/1,250,000 cc is shown by the bracket. The arrow shows a bacterium.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
If the bacteria are diluted, such as by mixing the bacteria with dye before being placed in the counting chamber, then this dilution must also be considered in the final calculations.
The formula used for the direct microscopic count is:
The number of bacteria per cc =
The average number of bacteria per large double-lined square X
The dilution factor of the large square (1,250,000) X
The dilution factor
of any dilutions made prior to placing the sample
in the counting chamber, e.g., mixing the bacteria with dye
C. TURBIDITY
As seen in Lab 2, when you mix the bacteria growing in a liquid medium, the culture appears turbid. This is because a bacterial culture acts as a colloidal suspension that blocks and reflects light passing through the culture. Within limits, the light absorbed by the bacterial suspension will be directly proportional to the concentration of cells in the culture. By measuring the amount of light absorbed by a bacterial suspension, one can estimate and compare the number of bacteria present.
The instrument used to measure turbidity is a spectrophotometer. (See Fig. 5.) It consists of a light source, a filter which allows only a single wavelength of light to pass through, the sample tube containing the bacterial suspension, and a photocell that compares the amount of light coming through the tube with the total light entering the tube.
Fig. 5: A Spectrophotometer
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
The ability of the culture to block the light can be expressed as either percent of light transmitted through the tube or the amount of light absorbed in the tube. (See Fig. 6.) The percent of light transmitted is inversely proportional to the bacterial concentration. (The greater the number of bacteria, the lower the percent light transmitted.) The absorbance, or optical density, is directly proportional to the cell concentration. (The greater the number of bacteria, the higher the absorbance.)
Fig. 6: Illustration of the Concept Behind a Spectrophotometer
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Turbidimetric measurement is often correlated with some other method of cell count, such as the direct microscopic method or the plate count. In this way, turbidity can be used as an indirect measurement of the cell count. For example:
1. Several dilutions can be made of a bacterial stock.
2. A Petroff-Hausser counter can then be used to perform a direct microscopic count on each dilution.
3. Then a spectrophotometer can be used to measure the absorbance of each dilution tube.
4. A standard curve comparing absorbance to the number of bacteria can be made by plotting absorbance versus the number of bacteria per cc. (See Fig. 7A.)
5. Once the standard curve is completed, any dilution tube of that organism can be placed in a spectrophotometer and its absorbance read. Once the absorbance is determined, the standard curve can be used to determine the corresponding number of bacteria per cc. (See Fig. 7B.)
Fig. 7A: A Standard Curve Plotting the Number of Bacteria per cc versus Absorbance, Step 1
Fig. 7B: Using a Standard Curve to Determine the Number of Bacteria per cc in a Sample by Measuring the Sample's Absorbance, Step 2
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
MATERIALS
6 tubes each containing 9.0 ml sterile saline, 3 plates of Trypticase Soy agar, 2 sterile 1.0 ml pipettes, pipette filler, turntable, bent glass rod, dish of alcohol
ORGANISM
Trypticase Soy broth culture of Escherichia coli
PROCEDURE
A. Plate Count (to be done in pairs)
Videos reviewing techniques used in this lab:
How to Read and Aseptically Handle a Sterile 1 milliliter (ml) Pipette
How to Pipette Using Aseptic Technique and use a Vortex Mixer
How to Obtain Single Colonies for Counting Using the Spin Plate Method
1. Take 6 dilution tubes, each containing 9.0 ml of sterile saline. Aseptically dilute 1.0 ml of a sample of E. coli as shown in Fig. 7 and described below.
Fig. 7: Plate Count Dilution Procedure Used in Today's Lab.
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
a. Remove a sterile 1.0 ml pipette from the bag. Do not touch the portion of the pipette that will go into the tubes and do not lay the pipette down. From the tip of the pipette to the "0" line is 1 ml; each numbered division (0.1, 0.2, etc.) represents 0.1 ml. (See Fig. 8.)
Fig. 8: A 1.0 Milliliter (ml) Pipette
The 1.0 ml pipette uses the blue pipette filler. The divisions are shown in the right-hand photograph.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
b. Insert the cotton-tipped end of the pipette into a blue 2 ml pipette filler.
c. Place the lip of the culture tube at the opening of the bacto-incinerator for 2-3 seconds, insert the pipette to the bottom of the flask, and withdraw 1.0 ml (up to the "0" line; see Fig. 8) of the sample by turning the filler knob towards you. Draw the sample up slowly so that it isn't accidentally drawn into the filler itself. Reflame and cap the sample. (See Fig. 9.)
Fig. 9: Using a Pipette to Remove Bacteria from a Culture Tube
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
d. Flame the first dilution tube and dispense the 1.0 ml of sample into the tube by turning the filler knob away from you. Draw the liquid up and down in the pipette several times to rinse the pipette and help mix. Reflame and cap the tube.
e. Mix the tube thoroughly by either holding the tube in one hand and vigorously tapping the bottom with the other hand or by using a vortex mixer. (See Fig. 10.) This is to assure an even distribution of the bacteria throughout the liquid.
Fig. 10: Using a Vortex Mixer to Mix Bacteria Throughout a Tube
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
f. Using the same procedure, aseptically withdraw 1.0 ml. (see Fig. 8 above) from the first dilution tube and dispense into the second dilution tube. Continue doing this from tube to tube, as shown in Fig. 7 above, until the dilution is completed. Discard the pipette in the used pipette container.
Your instructor will demonstrate these pipetting and mixing techniques.
2. Using a new 1.0 ml pipette, aseptically transfer 0.1 ml (see Fig. 8) from each of the last three dilution tubes onto the surface of the corresponding plates of trypticase soy agar as shown in Fig. 7 and Fig. 11. Note that since only 0.1 ml of the bacterial dilution rather than the desired 1.0 ml is placed on the plate, the actual dilution on the plate is 1/10 the dilution of the tube from which it came.
Fig. 11: Using a Pipette to Transfer Bacteria to an Agar Plate
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
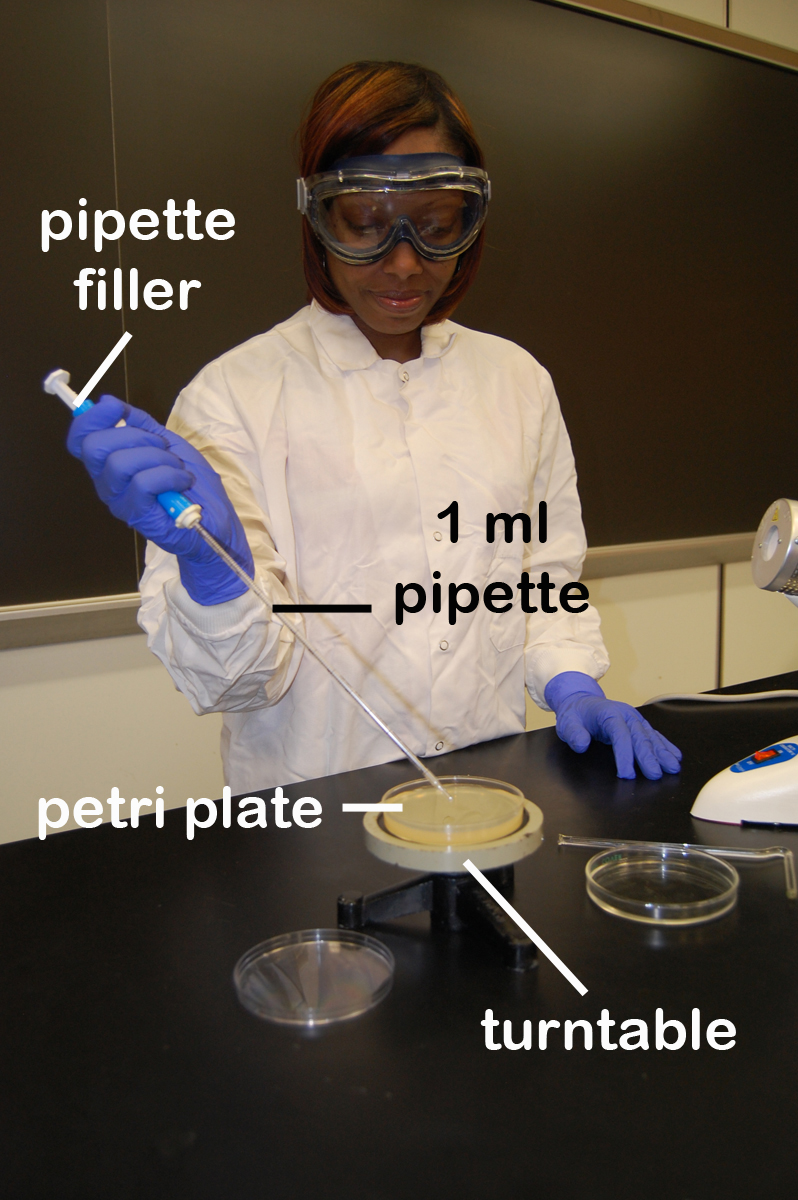
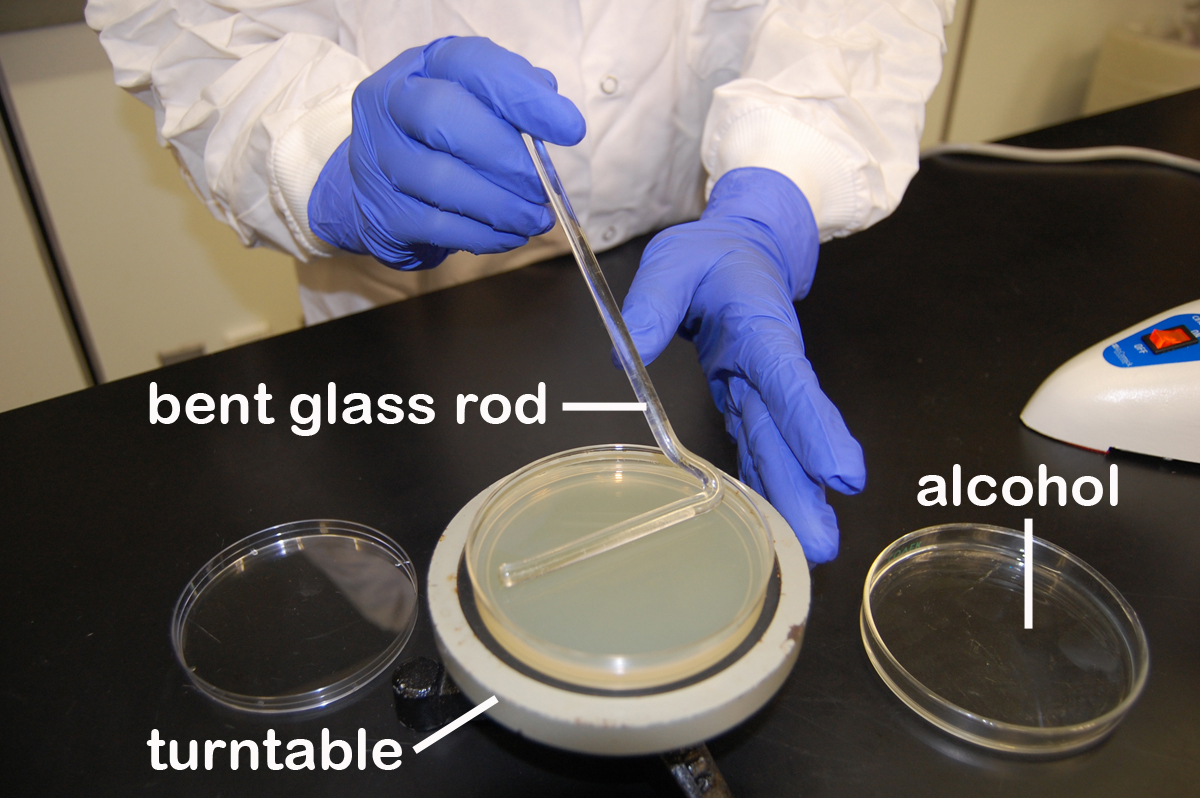
3. Using a turntable and sterile bent glass rod (see Fig. 12), immediately spread the solution over the surface of the plates as follows:
Fig. 12: Using a Bent Glass Rod and a Turntable to Spread a Bacterial Sample
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
a. Place the plate containing the 0.1 ml of dilution on a turntable.
b. Sterilize the glass rod by dipping the bent portion in a dish of alcohol and igniting the alcohol with the flame from your burner. Let the flame burn out.
c. Place the bent portion of the glass rod on the agar surface and spin the turntable for about 30 seconds to distribute the 0.1 ml of dilution evenly over the entire agar surface.
d. Replace the lid and resterilize the glass rod with alcohol and flaming.
e. Repeat for each plate.
f. Discard the pipette in the used pipette container.
4. Incubate the 3 agar plates upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section until the next lab period. Place the used dilution tubes in the disposal baskets in the hood.
The procedure is summarized in Figs. 13A-13I.
B. Direct Microscopic Method (demonstration)
1. Pipette 1.0 ml of the sample of E. coli into a tube containing 1.0 ml of the dye methylene blue. This gives a 1/2 dilution of the sample.
2. Using a Pasteur pipette, fill the chamber of a Petroff-Hausser counting chamber with this 1/2 dilution.
3. Place a coverslip over the chamber and focus on the squares using 400X (40X objective).
4. Count the number of bacteria in 5 large double-lined squares. For those organisms on the lines, count those on the left and upper lines but not those on the right and lower lines. Divide this total number by 5 to find the average number of bacteria per large square.
5. Calculate the number of bacteria per cc as follows:
The number of bacteria per cc =
The average number of bacteria per large square X
The dilution factor of the large square (1,250,000) X
The dilution factor of any dilutions made prior to placing the sample
in the counting chamber, such as mixing it with dye (2 in this case)
C. Turbidity
Your instructor will set up a spectrophotometer demonstration illustrating that as the number of bacteria in a broth culture increases, the absorbance increases (or the percent light transmitted decreases).
RESULTS
A. Plate Count
1. Choose a plate that appears to have between 30 and 300 colonies.
- Sample 1/100,000 dilution plate. (See Fig. 14A.)
- Sample 1/1,000,000 dilution plate. (See Fig. 14B.)
- Sample 1/10,000,000 dilution plate. (See Fig. 14C.)
Fig. 14A: 1/100,000 Dilution of Bacterium (10- 5)
Fig. 14B: 1/1,000,000 Dilution of Bacterium (10- 6)
Fig. 14C: 1/10,000,000 Dilution of Bacterium (10- 7)
This plate has over 300 colonies and cannot be used for counting.
This plate has between 30 and 300 colonies and is a suitable plate for counting.
This plate has less than 30 colonies and cannot be used for counting.Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
Gary E. Kaiser, Ph.D.
Professor of Microbiology
The Community College of Baltimore County, Catonsville Campus
This work is licensed under a Creative Commons Attribution 3.0 Unported License
2. Count the exact number of colonies on that plate using the colony counter (as demonstrated by your instructor).
3. Calculate the number of CFUs per ml of original sample as follows:
The number of CFUs per ml of sample =
The number of colonies (30-300 plate) X
The dilution factor of the plate counted
____________ = Number of colonies
____________ = Dilution factor of plate counted
____________ = Number of CFUs per ml
4. Record your results on the blackboard.
B. Direct Microscopic Method
Observe the demonstration of the Petroff-Hausser counting chamber.
C. Turbidity
Observe your instructor's demonstration of the spectrophotometer.
.
PERFORMANCE OBJECTIVES FOR LAB 4
After completing this lab, the student will be able to perform the following objectives:
DISCUSSION
1. State the formula for determining the number of CFUs per ml of sample when using the plate count technique.
2. When given a diagram of a plate count dilution and the number of colonies on the resulting plates, choose the correct plate for counting, determine the dilution factor of that plate, and calculate the number of CFUs per ml in the original sample.
- Plate count practice problems
3. State the principle behind the direct microscopic method of enumeration.
4. State the formula for determining the number of bacteria per cc of sample when using the direct microscopic method of enumeration.
5. When given the total number of bacteria counted in a Petroff-Hausser chamber, the total number of large squares counted, and the dilution of the bacteria placed in the chamber, calculate the total number of bacteria per cc in the original sample.
- Direct microscopic count practice problems
6. State the function of a spectrophotometer.
7. State the relationship between absorbance (optical density) and the number of bacteria in a broth sample.
8. State the relationship between percent light transmitted and the number of bacteria in a broth sample.
PROCEDURE
1. Perform a serial dilution of a bacterial sample according to instructions in the lab manual and plate out samples of each dilution using the spin-plate technique.
RESULTS
1. Count the number of colonies on a plate showing between 30 and 300 colonies and, by knowing the dilution of this plate, calculate the number of CFUs per ml in the original sample.
SELF-QUIZ
Microbiology Laboratory Manual by Gary E. Kaiser, PhD, Professor of Microbiology
is licensed under a Creative Commons Attribution 4.0 International License.
Last updated: March, 2023
Please send comments and inquiries to Dr. Gary Kaiser